Hi! I'm Tommy Tang
I am a computational biologist with six years of wet lab experience and over ten years of computation experience. I will help you to learn computational skills to tame astronomical data and derive insights. Check out the resources I offer below and sign up for my newsletter! https://github.com/crazyhottommy/getting-started-with-genomics-tools-and-resources
essential unix program every computational biologist needs and common mistakes when analyzing scRNAseq data
Hello Bioinformatics lovers, I can not believe it is almost the middle of 2024! I hope this year you have improved your computational skills. Remember, any skills need time to be good at. Essential Unix program you need Today, I am going to introduce you an essential Unix program called "screen". I am surprised that many experienced bioinformaticians do not know it. It is a program that can give you multiple windows in a terminal and keep your ssh connection persistent. If you do not...
2 days ago • 2 min read
Most neglected skill: organize your computational biology project
Hi Bioinformatics lovers, Welcome, all the new subscribers! Every Saturday you should expect to receive this short newsletter from me. Today, we will cover one important topic that is less taught: how to organize your computational biology project. If you prefer to watch videos, watch it here. I highly recommend you read this paper: A Quick Guide to Organizing Computational Biology Projects. For every project, you should have a consistent folder structure. You do not have to follow exactly...
9 days ago • 1 min read

do not repeat yourself. computers are good at repetitive work
Hi Bioinformatics lovers, I turned 38 this week. I typed my first "hello world" 12 years ago. I wrote it down in my first programming book: Python Programming for the Absolute Beginner. I have written a post on re-inventing yourself https://lnkd.in/eC_ChYst If you do not go out of your comfort zone, you will never grow.However, the growth will take time, sometimes taking a decade. But if you focus on doing small things consistently, you will see the success https://lnkd.in/eyuSRAGAIf you are...
23 days ago • 1 min read
what's the p-value and log2Fold change to use? Bioinformatics is not (just) statistics
Hello Bioinformatics lovers, I was asked this question very often: “Tommy, what’s the p-value cutoff should I use to determine the differentially expressed genes; what log2 Fold change cutoff should I use too?” For single-cell RNAseq quality control, what’s the cutoff for mitochondrial content? My answer is always: it depends. I was joking: determining a cutoff is 90% of the work a bioinformatician does. Why is that? read the full post here:...
30 days ago • 1 min read
Master these six types of plots, you can reproduce 90% of the figures in any genomics paper
Hello Bioinformatics lovers, I can not believe it is already the second quarter of 2024. Are you learning new things about computational biology? If you master these 6 types of plots: bar plots, scatter plots, line plots, histograms, boxplots/violin plots, and heat maps, you will be able to reproduce 90% of the figures in the genomics paper. Watch my video here. I will prove you at the end of the video that most of the figures are of those six basic types. The dotplot commonly used in...
about 1 month ago • 1 min read
How to learn computational biology the right way
Hello, Bioinformatics lovers, This is Tommy. Usually, you should receive my weekly newsletter on Saturday morning. I was away last week most of the time at Iowa State University https://idgp-biosci-symposium.sites.iastate.edu/2024-speaker-information for a symposium. I loved interacting with the students and seeing the awesome science they were doing. However, biology has become a data-intensive field. Lacking computational skills hinders their ability to analyze their data. Most of the talks...
about 1 month ago • 1 min read

can you determine the best resolution for scRNAseq clustering?
Hello Bioinformatics lovers, First of all, some encouragement. I found this old tweet from me 11 years ago. This is my audacious goal now: Ming Tommy Tang: On A Mission To Teach 1M People Bioinformatics The short answer is: yes, it is possible. You can achieve much more than you think you can! When I started learning 12 years ago, I had little clue. Now at least you have me. I am here to help! Let's dive into today's newsletter:) One of the core questions in scRNAseq is to choose the right...
about 2 months ago • 1 min read
Tutorial made for you: from fastq to differentially expressed genes
Hi Bioinformatics lovers, How is your weekend going? We had a leaking toilet, so it is a little hectic with three kids. Anyway, you should expect a newsletter from me every Saturday morning. Today is a little delayed. For those who want to learn bulk RNAseq analysis, check out these two tutorials: How to preprocess GEO bulk RNAseq data with salmon https://divingintogeneticsandgenomics.com/post/how-to-preprocess-geo-bulk-rnaseq-data-with-salmon/ Downstream of bulk RNAseq: read in salmon output...
about 2 months ago • 1 min read

Bioinformatics? You do not need machine learning most of the time
Hello Bioinformatics lovers, I know many of you want to use machine learning in bioinformatics. While it sounds exciting, the mundane work for bioinformatics is usually data cleaning, wrangling, and plotting. Mastering the basic skills of those can already get you very far in terms of solving practical bioinformatics problems. I am not saying machine learning is not important. I am also learning it and keeping myself in the front of deep learning. While the world is duped with more...
2 months ago • 1 min read
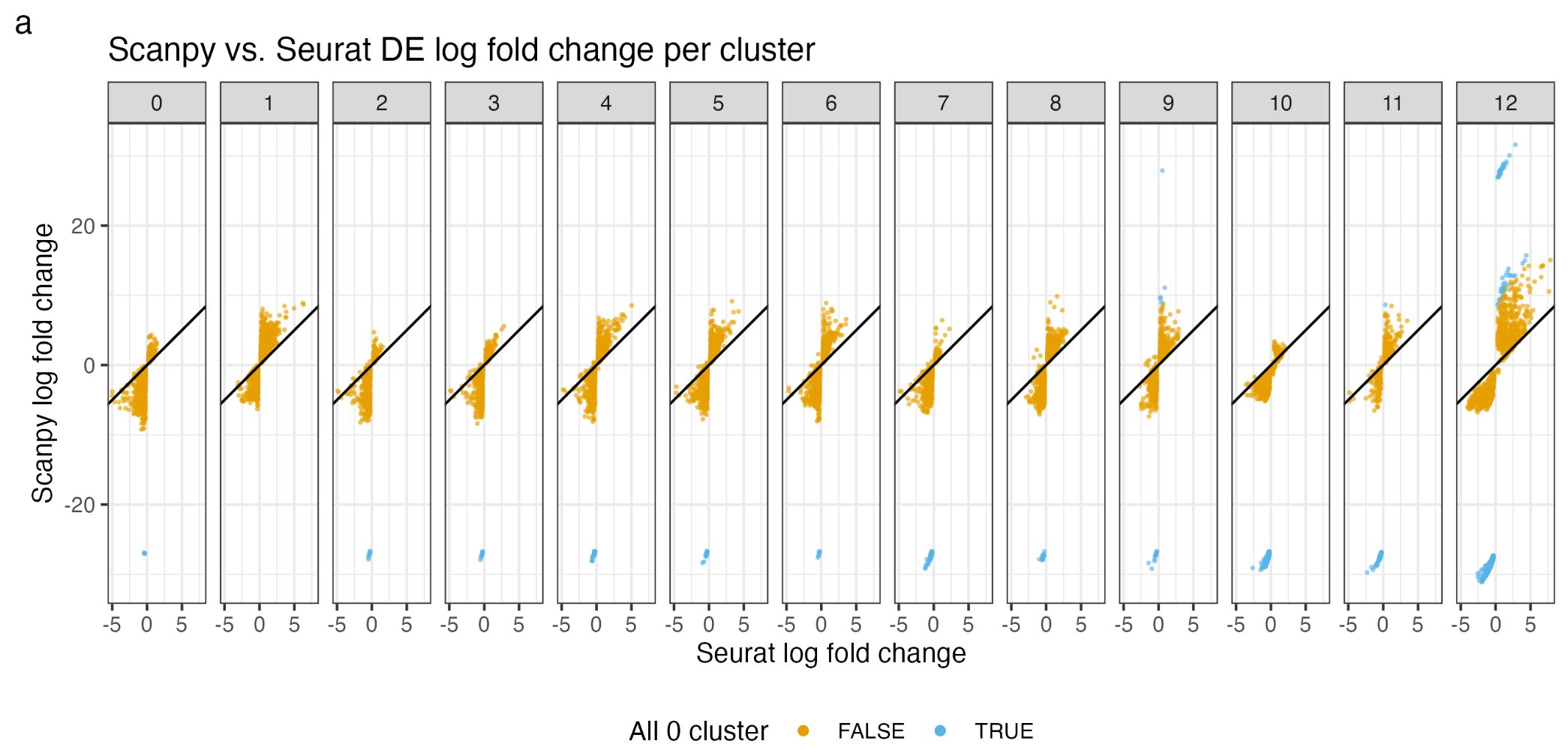
Seurat and Scanpy log2Fold change discrepancy for single cell data
Hello Bioinformatics lovers, If you work on single-cell RNAseq data, do you know Seurat (in R) and scanpy (in python) had a discrepancy in calculating log2Fold change for marker genes per cluster? Read more details in this blog post: https://divingintogeneticsandgenomics.com/post/do-you-really-understand-log2fold-change-in-single-cell-rnaseq-data/ In this video, I show you how to use google gemini multi-modal to write math formula in Latex by feeding a screenshot to it: Other useful resources...
2 months ago • 1 min read
I am a computational biologist with six years of wet lab experience and over ten years of computation experience. I will help you to learn computational skills to tame astronomical data and derive insights. Check out the resources I offer below and sign up for my newsletter! https://github.com/crazyhottommy/getting-started-with-genomics-tools-and-resources