I am a computational biologist with six years of wet lab experience and over ten years of computation experience. I will help you to learn computational skills to tame astronomical data and derive insights. Check out the resources I offer below and sign up for my newsletter! https://github.com/crazyhottommy/getting-started-with-genomics-tools-and-resources
Seurat and Scanpy log2Fold change discrepancy for single cell data
|
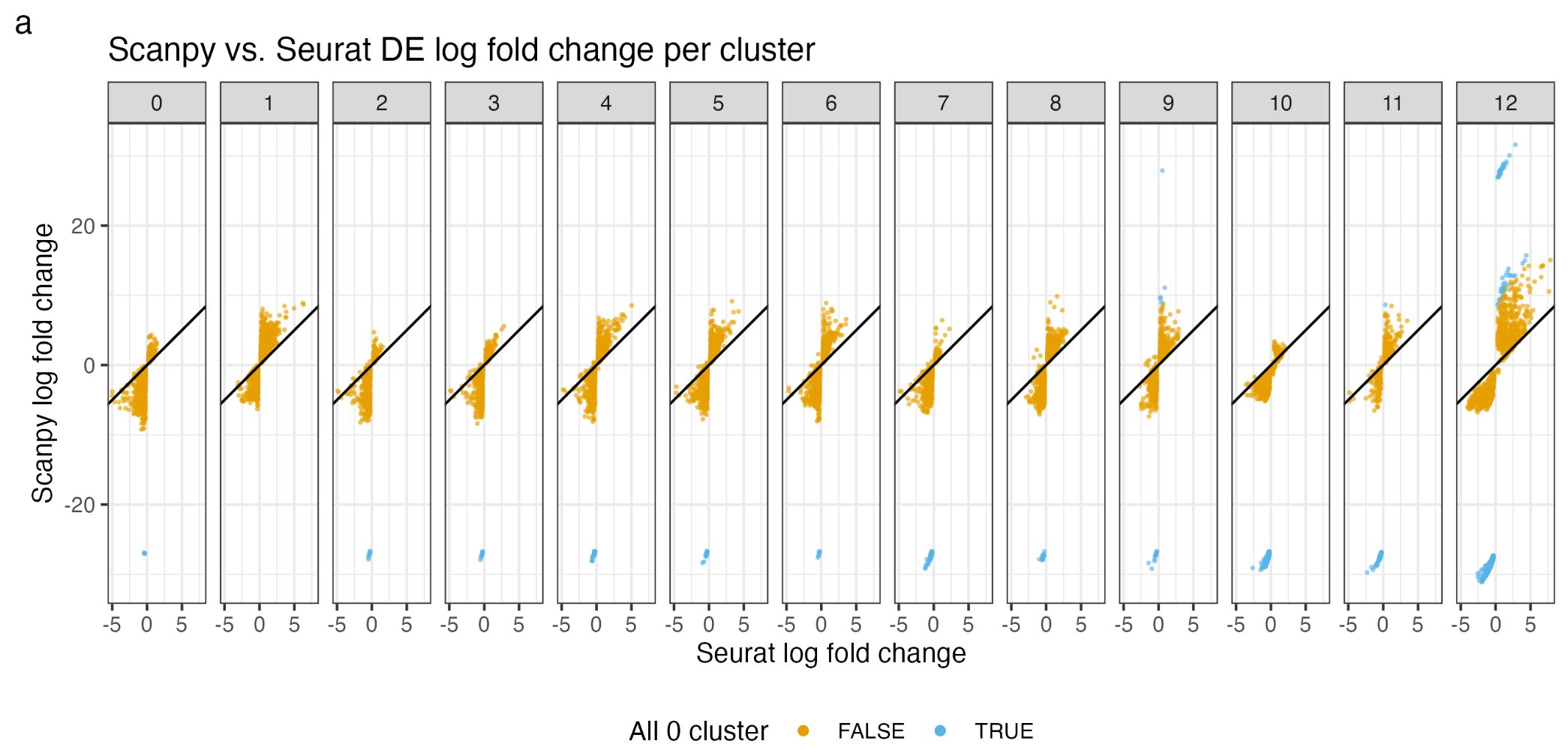
Hello Bioinformatics lovers, If you work on single-cell RNAseq data, do you know Seurat (in R) and scanpy (in python) had a discrepancy in calculating log2Fold change for marker genes per cluster?
Read more details in this blog post: https://divingintogeneticsandgenomics.com/post/do-you-really-understand-log2fold-change-in-single-cell-rnaseq-data/ In this video, I show you how to use google gemini multi-modal to write math formula in Latex by feeding a screenshot to it: Other useful resources this week:
Happy Learning! Tommy Let's connect on twitter and Linkedin! |
Hi! I'm Tommy Tang
I am a computational biologist with six years of wet lab experience and over ten years of computation experience. I will help you to learn computational skills to tame astronomical data and derive insights. Check out the resources I offer below and sign up for my newsletter! https://github.com/crazyhottommy/getting-started-with-genomics-tools-and-resources